Pig Genome Functional Annotation: Unlocking Secrets of Complex Traits and Human Disease
Method of comparative genomics, 60’s to recently
Pig Genome Functional Annotation: Unlocking Secrets of Complex Traits and Human Disease
The functional annotation of the pig genome, meticulously explored in the groundbreaking study "Pig genome functional annotation enhances the biological interpretation of complex traits and human disease," represents a watershed moment in our understanding of both livestock biology and human health. By meticulously integrating 223 epigenomic and transcriptomic datasets spanning 14 critical tissues, the authors unveil a treasure trove of regulatory elements, shed light on the dynamic epigenetic landscape, and forge connections between genetic variations and complex traits.
This landmark study promises to revolutionize research in diverse domains, from improving agricultural practices to advancing personalized medicine.
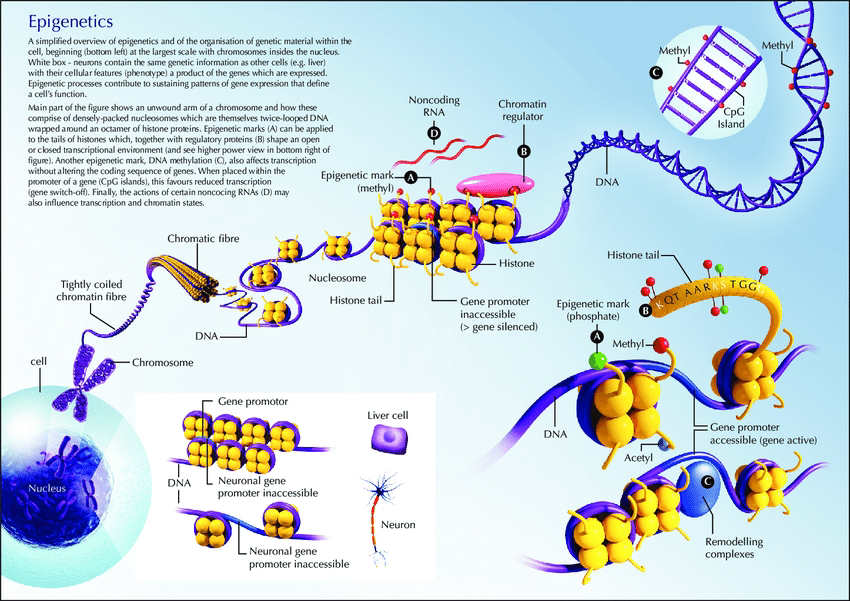
Delving into the Epigenetic Landscape: A Symphony of Chromatin States
The study meticulously delves into the pig's epigenetic landscape, uncovering the orchestra of chromatin states that regulate gene expression across various tissues. By functionally annotating 15 distinct chromatin states, the authors map their tissue-specific regulatory activities, providing a nuanced understanding of how DNA modifications orchestrate gene expression variations. This intricate tapestry of epigenetic marks offers invaluable insights into gene regulation, potentially guiding the development of targeted interventions for manipulating gene expression pathways in pigs and potentially other organisms.
Linking Variants to Complex Traits: Demystifying Disease Mechanisms
One of the study's most captivating aspects is its exploration of how genetic variants can influence complex traits in pigs. By demonstrating that variations associated with these traits are significantly enriched in active promoters and enhancers, the authors establish a crucial link between specific DNA sequences and complex biological phenotypes. This knowledge empowers researchers to pinpoint the genetic underpinnings of traits of economic importance in pigs, such as meat quality, growth rate, and disease resistance. Moreover, it sheds light on the regulatory mechanisms governing similar traits in humans, paving the way for the development of novel diagnostic and therapeutic tools for complex human diseases.
Evolutionary Footprints: Unraveling Domestication and Adaptation
Intriguingly, the study delves into the evolutionary forces that have shaped the pig genome during domestication and adaptation. The authors reveal distinct tissue-specific regulatory selection signatures between Asian and European pig breeds, highlighting how selective pressures have differentially driven regulatory evolution in different populations. This finding unveils the nuanced genetic and phenotypic consequences of diverse domestication histories, offering valuable insights into the process of breed development and adaptation in other organisms as well.
Pig vs. Mouse: Choosing the Right Model for Human Disease
By meticulously comparing porcine functional annotations with data from human and mouse models, the study provides invaluable guidance on selecting the most suitable model organism for studying specific human diseases. The authors demonstrate remarkable conservation of tissue-specific epigenetic signatures across species, suggesting that pigs could be more appropriate models than mice depending on the specific disease under investigation. This crucial understanding has the potential to significantly expedite advancements in human disease research, leading to more relevant and efficacious treatment strategies.
Future Horizons: Translational Potential and Ethical Considerations
The comprehensive understanding gleaned from this study unlocks a kaleidoscope of future possibilities. The detailed characterization of regulatory elements could guide the development of gene-editing tools for enhancing desirable traits in pigs, thereby revolutionizing agricultural practices. Furthermore, the elucidation of shared regulatory mechanisms between pigs and humans opens doors for investigating human diseases in a more physiologically relevant context, potentially leading to personalized medicine breakthroughs.
Conclusion: A Pivotal Step Towards a Healthier Future
The meticulous functional annotation of the pig genome stands as a testament to the power of collaborative research and innovative methodologies. By unveiling the intricate interplay between regulatory elements, genetic variations, and complex traits, this study paves the way for significant advancements in pig breeding, human disease research, and personalized medicine. As we move forward, prioritizing ethical considerations and open dialogue will be essential in harnessing the full potential of this groundbreaking work to create a healthier future for humans and animals alike.
Pig's Puzzle Pieces: Unlocking Genomes, Epigenetics, and Complex Traits
The article sheds light on how studying pigs can contribute to understanding human health and evolution. It highlights the importance of epigenetics, challenging the traditional "gene-centric" view of comparative genomics.
The research delves into the pig's genome, creating a dynamic picture of the epigenetic landscape. This landscape refers to the chemical modifications that influence gene expression without altering the DNA sequence itself.
Here's where the old view gets shaken up:
Genes aren't the sole players: Traditionally, geneticists focused on identifying genes associated with traits. This study emphasizes the crucial role of regulatory elements (promoters and enhancers) in controlling gene expression. These elements are often influenced by epigenetic modifications.
Beyond the sequence: DNA sequence alone doesn't hold all the answers. Counting nucleotides alone gives preposterous comparisons.
Epigenetic marks like DNA methylation and histone modifications play a significant role in shaping gene activity and complex traits.
Tissue-specific insights: The study reveals how epigenetic marks differ across tissues, highlighting the importance of considering tissue-specific regulatory mechanisms.
The research also demonstrates how genetic variants associated with complex traits and pig evolution are enriched in these active regulatory elements. This opens doors to understanding the molecular mechanisms underlying important features like growth, fat deposition, and disease resistance.
Furthermore, the study compares the pig's regulatory elements with human and mouse counterparts. This reveals interesting insights into the conservation of epigenetic signatures across species, suggesting which animal model might be more suitable for studying different human diseases.
In essence, this research emphasizes the power of integrating functional annotations and epigenomics into understanding complex traits and diseases. By looking beyond genes and incorporating the dynamic epigenetic layer, we gain a deeper understanding of how organisms function and evolve. The pig, once considered a simple livestock animal, now emerges as a valuable model for biomedical research, challenging the traditional gene-centric view and opening new avenues for scientific exploration.
Snippets:
Pig genome functional annotation enhances the biological interpretation of complex traits and human disease.
The functional annotation of livestock genomes is crucial for understanding the molecular mechanisms that underpin complex traits of economic importance, adaptive evolution and comparative genomics and epigenomics.
We systematically describe the dynamic epigenetic landscape across tissues.
We demonstrate that genomic variants associated with complex traits and adaptive evolution in pig are significantly enriched in active promoters and enhancers (epigenetics).
Compared with human and mouse epigenomes, we show that porcine regulatory elements are more conserved in DNA sequence, under both rapid and slow evolution, than those under neutral evolution across pig, mouse, and human.
We demonstrate that, depending on the traits, mouse or pig might be more appropriate biomedical models for different complex traits and diseases.
Comparative analysis of epigenomes and transcriptomes across species could provide novel insights into the molecular mechanisms underlying human disease.
For understanding some human diseases. However, compared with the mouse, pig (Sus scrofa) has more anatomical and physiological similarities to humans.
Following ENCODE (epigenetics) and Roadmap Epigenomics projects, the Functional Annotation of Animal Genomes (FAANG) initiative, although still in its infancy, has made great progress towards annotating functional elements in many tissues across multiple domestic species, including pigs.
We find that tissue-specific regulatory elements were enriched for the potential causative variants of complex phenotypes by integrating a variety of large-scale genome-wide association studies (GWAS) and expression quantitative trait loci (eQTL)(epigenetics).
We show that tissue-specific regulatory elements (epigenetics) likely played important roles in pig domestication.
These comparisons demonstrate conservation of tissue-specific epigenetic signatures, suggesting that, depending on the specific human diseases under investigation, either the pig or the mouse may be a more suitable animal model.
Our results demonstrated that variants of complex traits and eQTLs of growth-related traits were significantly enriched in the active promoters and enhancers annotated by this study.
Our analysis illustrated that signatures of domestication were significantly enriched in porcine regulatory elements.
FAANG Consortium, more epigenomic data will be available from diverse samples, such as reproductive tissues, additional developmental stages, and different physiological states.
This atlas of functional elements provided a unique opportunity for comparative epigenomic analysis between human, mouse and pig, the results of which can inform which species constitute the most appropriate biomedical model(s) for specific human diseases.
confirming the hypothesis that elasticity of regulatory conservation (epigenetics) may play an important role in the evolution of the less conserved regions (impact of negative selection pressure).
human-specific promoters in brain tissues were enriched in intelligence-related genes, which suggests a critical role for epigenomic regulation of novel biological function in humans.
different mammalian species. Although the findings from our study are intriguing, experimental studies and more epigenomic data from additional tissues, cell types, and species – such as non-human primates – will be needed to extend and functionally validate the biological mechanisms that underpin complex traits and diseases.






Comments
Post a Comment